Failing
ugropy may fail to obtain the subgroups of a molecule for a certain model for two reasons: either there is a bug in the code, or the molecule cannot be represented by the subgroups of the failing model.
ugropy utilizes SMARTS for the representation of functional groups to inquire whether the molecule contains those structures. Let’s examine the functional group list for the classic liquid-vapor UNIFAC model.
[1]:
try:
import google.colab
IN_COLAB = True
except:
IN_COLAB = False
if IN_COLAB:
%pip install ugropy
[2]:
from ugropy import unifac
unifac.subgroups
[2]:
| smarts | molecular_weight | |
|---|---|---|
| group | ||
| CH3 | [CX4H3] | 15.03500 |
| CH2 | [CX4H2] | 14.02700 |
| CH | [CX4H] | 13.01900 |
| C | [CX4H0] | 12.01100 |
| CH2=CH | [CH2]=[CH] | 27.04600 |
| ... | ... | ... |
| NCO | [NX2H0]=[CX2H0]=[OX1H0] | 42.01700 |
| (CH2)2SU | [CH2]S(=O)(=O)[CH2] | 92.11620 |
| CH2CHSU | [CH2]S(=O)(=O)[CH] | 91.10840 |
| IMIDAZOL | [c]1:[c]:[n]:[c]:[n]:1 | 68.07820 |
| BTI | C(F)(F)(F)S(=O)(=O)[N-]S(=O)(=O)C(F)(F)F | 279.91784 |
113 rows × 2 columns
For example, let’s check the SMARTS representation of the alcohol group ACOH:
[3]:
unifac.subgroups.loc["ACOH", "smarts"]
[3]:
'[cH0][OH]'
The SMARTS representation it’s telling us that the OH group it’s, of course, a hydroxyl group bounded by a single bound to an aromatic carbon atom.
An example of a molecule that cannot be represented by UNIFAC groups:
[4]:
from rdkit.Chem import Draw
mol = unifac.get_groups("C1(=CC=CC=C1)OC(C)(C)C", "smiles")
Draw.MolToImage(mol.molecule)
[4]:
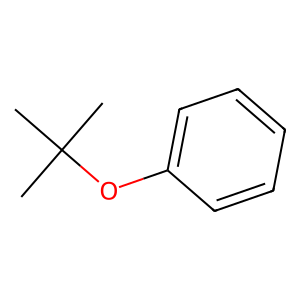
[5]:
print(mol.subgroups)
{}
The library “fails” to obtain any functional groups to accurately represent the molecule. This failure is represented by an empty dictionary. In this case, the “fail” is correct, but it could fail due to errors in the groups SMARTS representations or the algorithm, resulting in an empty dictionary as well. Currently, the supported models are tested against 444 different molecules.
If you encounter a failing representation, you can examine the structure of the molecule and the list of functional groups of the failing model. If you determine that the molecule can indeed be modeled, you may have discovered a bug. Feel free to report the issue on the repository along with the failing molecule’s SMILES/name, the failing model and the ugropy version.
More than one solution
Models like UNIFAC or PSRK can have multiple solutions to represent a molecule, and ugropy tries its best to find them all. In such cases, you will receive a list of dictionaries, each containing one of the solutions found. Let’s take a look.
[6]:
from ugropy import unifac
from rdkit.Chem import Draw
mol = unifac.get_groups("CCCC1=CC=C(CC(=O)OC)C=C1", "smiles", search_multiple_solutions=True)
Draw.MolToImage(mol[0].molecule, highlightAtoms=[7])
[6]:
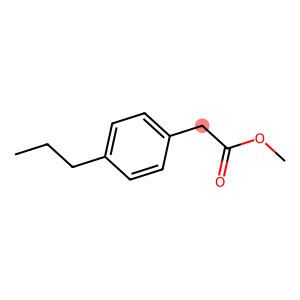
This molecule can be modeled in two ways depending on how we treat the CH2 attached to the ring and the ester carbon (highlighted in red). We can either form an ACCH2 group and model the ester group with COO, or we can use an AC group and model the ester group with CH2COO.
[7]:
print("Solution 1:")
print(mol[0].subgroups)
print("solution 2:")
print(mol[1].subgroups)
Solution 1:
{'CH3': 2, 'CH2': 1, 'ACH': 4, 'ACCH2': 2, 'COO': 1}
solution 2:
{'CH3': 2, 'CH2': 1, 'ACH': 4, 'AC': 1, 'ACCH2': 1, 'CH2COO': 1}
[8]:
mol[0].draw(width=800)
[8]:
[9]:
mol[1].draw(width=800)
[9]:
This could be useful in cases where some groups have more interaction parameters than others in the mixture that you want to model with UNIFAC. Alternatively, you can try both approaches and compare if there are any differences.